New microchip enables faster DNA separation
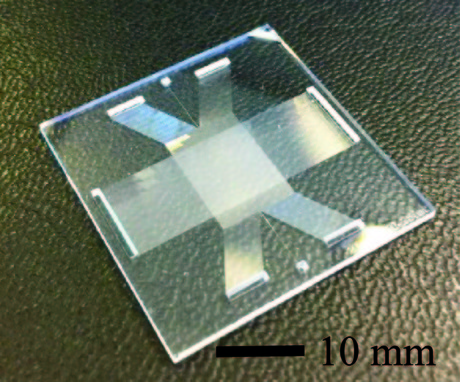
A new glass micochip, developed by researchers from the University of Twente (UT) in the Netherlands, enables faster separation and purification of DNA fragments.
The new chip is capable of fractionating DNA fragments within just a few minutes, while conventional approaches take hours, according to the UT. The chip does this in high resolution and also purifies the fragments; it removes the other salts in the DNA sample. Tiny amounts of DNA, like in medical diagnostics or in forensics, will be sufficient. The fragments are typically in the size range of second-generation DNA sequencing, the next step after the well-known Human Genome Project.
UT scientist Burcu Gumuscu and her colleagues achieved this high speed and resolution by inventing a new approach to the common technique of gel electrophoresis. In the conventional approach, the DNA moves in a gel by an applied electric field. Larger fragments will move slower than small ones, and in this way size separation is possible. But the electric field has one direction and the molecules move in a straight line. What about varying the field, the UT scientists thought. If you periodically apply an electric field of different magnitude in the perpendicular direction, the fragments will also respond to this. They don’t just move in a straight line now, but somewhere in between the two field directions. As large fragments respond differently — like a snake moves — to the fields than small ones, fragments can be separated.
For collecting the fragments, the chip has arrays of micro channels on the sides of a square separation chamber of about 1 square centimetre. Next to that, it has a DNA reservoir and electrodes for applying the electric fields. The chip is said to be relatively easy to produce using fundamental cleanroom techniques of the MESA+ NanoLab, and cheap. It is versatile as well, as the type and concentration of gel and the electric fields can be adjusted to the application. Protein separation is one of the other possible application areas.
The research shows that DNA fragments between 0.5 and 10 kbp (1 kbp = 1000 base pairs) can be fractionated within two minutes, at high resolution. These are typically the smaller fragments, essential for the second-generation DNA sequencing.
The UT scientists published their results in Microsystems and Nano Engineering, a Nature publication.
A pre-emptive approach to treating leukaemia relapse
The monitoring of measurable residual disease (MRD), medication and low-dose chemotherapy is...
Long COVID abnormalities appear to resolve over time
Researchers at UNSW's Kirby Institute have shown that biomarkers in long COVID patients have...
RNA-targeted therapy shows promise for childhood dementia
Scientists have shown that a new RNA-targeted therapy can halt the progression of a specific type...







