Over 10,000 proteins in cancer cells identified
Researchers at Technische Universität München (TUM) have conducted what is being called the biggest study into cancer cells to date. They have decoded the proteome of 59 tumour cell lines from the US National Cancer Institute, known as the ‘NCI-60’ cell lines, which represent the most common tumour diseases in nine tissues (eg, brain, breast, bowels, skin, blood).
Previously, researchers were primarily interested in mutations in DNA, which is the blueprint for proteins. But as Dr Amin Gholami notes, “... it is proteins that make the difference between a healthy cell and a tumour cell”, yet before now “there was little knowledge on the proteome of the NCI-60 cell lines”. The identification of the proteome - “the protein portfolio of tumour cells”, according to Professor Bernhard Küster - is expected to assist in finding new drug targets, since “nearly all anti-tumour drugs are targeted against cellular proteins”, he adds.
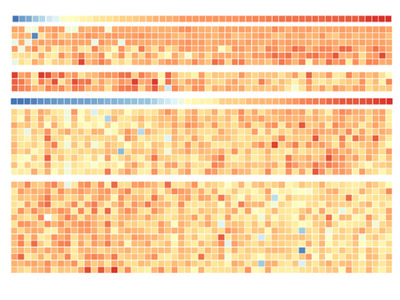
The scientists found 10,350 different proteins in the cell lines, 5578 of which are present in varying abundance in tumour cell lines of all types of tissue and thus assumed by Professor Küster to be “the core proteome of cancer”. The researchers also found some clearly distinguishable protein profiles between the tumour cells of different tissues - a clear indication that the tumour cells possess typical characteristics of healthy cells of the same tissue.
Meanwhile, 375 of the proteins belong to the kinase group, responsible for transmitting signals which cells use to control, for example, their metabolic reactions, their division and their communication with other cells in the tissue. Dr Zhixiang Wu explains, “Unlike healthy cells, tumour cells are open to many signalling pathways in order to activate their division and ensure their survival.” Professor Küster added that the kinases “play a part in the continuous reproduction process of tumour cells”.
The researchers also demonstrated that the protein pattern of the cells determines the effectiveness of cancer drugs. Dr Hannes Hahne said the team “investigated how the cell lines react to 108 different cancer drugs and found that there are indeed proteins that indicate whether a cell will respond to certain therapeutic agents - or whether it will be resistant to the drug”. Some of these protein markers were already known, while others could potentially be used in future.
The researchers say they “anticipate that proteomics will play an increasing role in molecular profiling of cancer”. They have published their results in the journal Cells Reports, as well as in an online database which can be accessed here.
Blood-based biomarker can detect sleep deprivation
The biomarker detected whether individuals had been awake for 24 hours with a 99.2% probability...
Epigenetic signature helps to diagnose rare breast tumour
The current way of diagnosing phyllodes tumours is to analyse their cellular features under a...
New instrument measures cardiovascular disease biomarkers
CVD-21 enables a 'liquid cardiovascular biopsy' for quantification of multiple...







