3D imaging a cancer protein
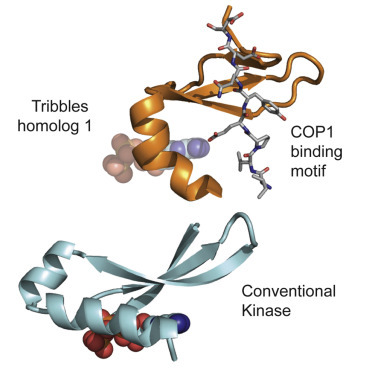
Scientists at the Walter and Eliza Hall Institute (WEHI) have created what is said to be the first 3D image of a key protein known to be involved in the development of blood and other cancers. The results of their study have been published in the journal Structure.
The protein TRIB1 was characterised by WEHI researchers Dr James Murphy and Dr Isabelle Lucet in collaboration with Dr Peter Mace from the University of Otago, New Zealand. TRIB1 is part of the protein family Tribbles, which play diverse roles in cell signalling and development. Excess Tribbles drive the abnormal production of immune cells, causing a type of blood cancer called acute myeloid leukaemia (AML).
TRIB1 plays a vital role in controlling how and when other proteins are degraded, which is essential for managing protein levels in the cell. As explained by Dr Murphy, “The amount of protein in a cell depends on the balance between production and degradation. Defects in protein degradation, or in the controllers of protein degradation, disrupt this balance and can lead to diseases such as cancer.”
Dr Murphy said TRIB1 is an unusual type of protein called a pseudokinase — one which has apparently lost its old activity and gained other functions. Using the Australian Synchrotron, the researchers were able to obtain detailed 3D images of TRIB1 and learn exactly how it developed its new functions.
“The powerful X-ray beams created by the synchrotron enabled us to see that TRIB1 has undergone huge contortions compared to its ancestors,” Dr Murphy said. “These structural changes prevent TRIB1 from driving chemical reactions and instead allow it to act as a scaffold to bring proteins together.
“The structure of TRIB1 is really exciting,” he added. “We can now see how TRIB1 is able to trigger protein destruction, which will provide critical clues for developing drugs that target TRIB1 to treat cancers.”
Dr Mace, who acted as lead investigator on the study, said the research could “help us design novel therapeutic agents for the treatment of AML”.
“For example, some AML patients have too much TRIB1, which causes a loss of proteins that would normally inhibit cancer,” he said. “Understanding the structure of TRIB1 provides critical clues about how we could block Tribbles for the treatment of AML.”
Omega-6 fatty acid could reduce your risk of bipolar
Arachidonic acid — a polyunsaturated omega-6 fatty acid — may exert an effect on...
Blood-based biomarker can detect sleep deprivation
The biomarker detected whether individuals had been awake for 24 hours with a 99.2% probability...
Epigenetic signature helps to diagnose rare breast tumour
The current way of diagnosing phyllodes tumours is to analyse their cellular features under a...







