Identifying new drug candidates using genome sequences
Scientists from The Scripps Research Institute (TSRI) have developed a new approach to designing drugs using genome sequences. The technique, described in the journal Nature Chemical Biology, was dubbed Inforna.
The study was led by Dr Matthew Disney, an associate professor at TSRI. Dr Disney and his team were already interested in understanding the binding of drugs to RNA folds and in manipulating microRNAs. Some microRNAs have been associated with diseases; miR-96, for example, is thought to promote cancer by discouraging a process called apoptosis, or programmed cell death, that can rid the body of cells that begin to grow out of control.
The Disney lab developed computational approaches that can mine information against genome sequences and all cellular RNAs with the goal of identifying drugs that target such disease-associated RNAs while leaving others unaffected. In their latest study, the Inforna technique identified optimal drug targets by mining a database of drug-RNA sequence (‘motif’) interactions against thousands of cellular RNA sequences.
“This is the first time therapeutic small molecules have been rationally designed from only an RNA sequence - something many doubted could be done,” said Dr Disney.
Using Inforna, the team identified compounds that can target microRNA-96, as well as additional compounds that target nearly two dozen other disease-associated microRNAs. They showed that the drug candidate that inhibited microRNA-96 also inhibited cancer cell growth. Furthermore, cells without functioning microRNA-96 were unaffected by the drug.
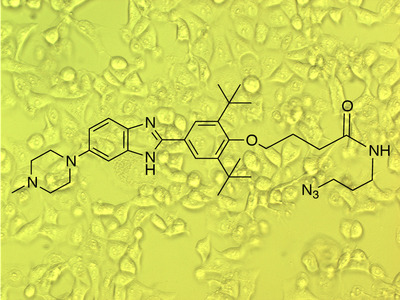
“This illustrates an unparalleled selectivity for the compound,” Dr Disney noted. “In contrast, typical cancer therapeutics target cells indiscriminately, often leading to side effects that can make these drugs difficult for patients to tolerate.”
He added that the new drug candidate, which is easy to produce and cell permeable, targets microRNA-96 far more specifically than the method to target RNA (using oligonucleotides) currently in use.
“With our program, we can identify compounds with high specificity,” said Sai Pradeep Velagapudi, the first author of the study and a graduate student working in the Disney lab. “In the future, we hope we can design drug candidates for other cancers or for any pathological RNA.”
Blood test could be used to diagnose Parkinson's earlier
Researchers have developed a new method that requires only a blood draw, offering a non-invasive...
Cord blood test could predict a baby's risk of type 2 diabetes
By analysing the DNA in cord blood from babies born to mothers with gestational diabetes,...
DNA analysis device built with a basic 3D printer
The Do-It-Yourself Nucleic Acid Fluorometer, or DIYNAFLUOR, is a portable device that measures...



