Creating 3D images with super-resolution microscopy
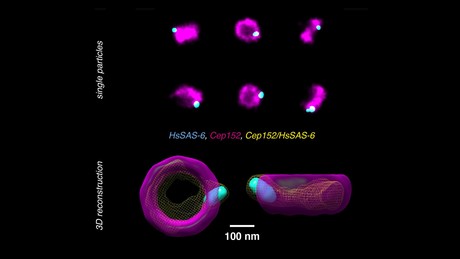
A new technique developed by Switzerland’s École Polytechnique Fédérale de Lausanne (EPFL) overcomes the noise and colour limitations of super-resolution microscopy by creating three-dimensional reconstructions from single-colour, two-dimensional images of protein complexes.
Super-resolution microscopy is a technique that can ‘see’ beyond the diffraction limit of light, exploiting fluorescence to allow scientists to observe cells and their interior structures and organelles in a way never before possible.
The disadvantage is that many of the molecular complexes inside cells are made up of multiple proteins — and since current techniques of super-resolution microscopy typically can only use one or two fluorescent colours, it is difficult to observe different proteins and decipher the complex architecture and underlying assembly mechanisms of the cell’s interior structures. An even greater challenge is to overcome the noise inherent to the super-resolution methods and fluorescent labelling, to achieve the full resolution potential.
Now, EPFL scientists have solved both problems by developing a new method to analyse and reconstruct super-resolution images and realign them in a way that multiple proteins can be placed within a single 3D volume. The method works with images taken with large field-of-view super-resolution microscopy, with each image containing hundreds of two-dimensional projections of a labelled structure in parallel.
Each 2D view represents a slightly different orientation of the structure, so that with a dataset of thousands of views, the method can computationally reconstruct and align the 2D images into a 3D volume. By combining information from a large number of single images, the noise is reduced and the effective resolution of the 3D reconstruction is enhanced.
The researchers tested their method on human centriole complexes — pairs of cylindrical molecular assemblies that are crucial in helping the cell divide — with the results published in the journal Nature Methods. They were able to uncover the 3D architecture of four proteins critical for centriolar assembly during organelle biogenesis, and believe their new approach allows for unlimited multiplexing capabilities.
“With this method, if the proteins in the structure can be labelled, there is no limit to the number of colours in the 3D reconstruction,” said Suliana Manley, whose lab headed the project. “Plus, the reconstruction is independent of the super-resolution method used, so we expect this analysis method and software to be of broad interest.”
A non-destructive way to locate microplastics in body tissue
Currently available analytical methods either destroy tissue in the body or do not allow...
Rapid imaging method shows how medicine moves beneath the skin
Researchers have developed a rapid imaging technique that allows them to visualise, within...
Fluorescent molecules glow in water, enhancing cell imaging
Researchers have developed a new family of fluorescent molecules that glow in a surprising way,...



