Minimising challenges in biological modelling
Thursday, 01 December, 2011
Simulation streamlines modelling workflow and provides one platform for model creation, simulation and analysis.
Modelling has long played a key role in the drug discovery process, from isolating drug targets to screening and pharmacokinetics. Many pharmaceutical companies are currently facing patent expirations without replacement candidates in the pipeline, or are struggling with high candidate attrition. Leaner pipelines results in necessary cost decreases, which can often lead to staff reductions. These struggles underscore the need for successful modelling in pharmaceutical R&D.
As a result of this increase in intensity and in application, the needs of pharma modellers have also drastically increased. In general, pharma modellers face two main tasks: first, to complete their projects quickly and second, to communicate their results to their peers in the scientific community and to corporate managers.
Modellers spend a great deal of time integrating various tools into their workflow to create a piecemeal solution that fits their needs. They use one tool to model data, another to perform simulation, and perhaps yet another tool for analysis. Once they have results, researchers will spend an even larger amount of time transferring the results into a format that scientists or executives will understand. This workflow is inefficient and an expensive waste of time.
SimBiology software was driven by customer demand to provide a modelling environment that minimises this inefficiency. Modellers can perform their entire project workflow in a single environment, from creating a model to simulation and analysis. The interface is graphical so modellers have a pathway diagram to easily communicate with scientists and management, yet the underlying technology is mathematically strong to meet modellers’ needs.
Method of operation
SimBiology offers several different modes of interaction, all of which are fully integrated. The SimBiology desktop is a customisable graphical user interface (GUI) that offers a point-and-click environment for modelling. Modellers can move windows around the desktop using drag and drop or menu selections.
Within the desktop, the block diagram editor allows you to work with the graphical view of your model in a drag-and-drop environment. This includes a library of predefined blocks you can use to create models. Lastly, for modellers who prefer to automate their modelling and analysis, all functionality can also be accessed from the command line.
Constructing models
SimBiology models consist of compartments, species, parameters, reactions, events, rules, kinetic laws and units. Users are presented with a variety of ways to create models such as dragging and dropping predefined blocks in the block diagram editor, entering equations and relevant parameters into the GUI, or by importing SBML files. Once a model has been created, you can alter the appearance of graphical entities, including replacing the entity with an image, to better visually communicate or organise your model as shown in Figure 1. Any altered entity can be saved into a library for future re-use. You can also add annotations, such as references or notes, to any entity by double-clicking on the element in the diagram editor.
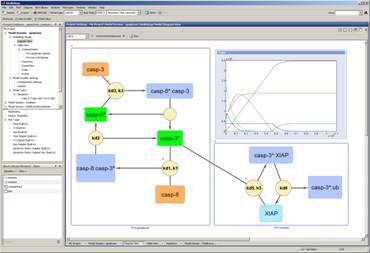
Figure 1: The block diagram editor lets users drag and drop models and elements.
Kinetic laws and units characterise the dynamics of the model. Kinetic laws and units are presented as built-in libraries or user-defined laws, and you can also add units. Variants, or alternate values for parameters, are applied to the model if you want to study various values for model components without creating a new model.
The output or response of a model, such as concentration as a function of time, can be determined by simulating the model with either stochastic or deterministic solvers. Results display in a figure window or can be embedded into the block diagram editor as shown in Figure 1. Simulation events can be applied to the model to define a sudden change in model behaviour such as a bolus injection given to a patient.
Analysis
Once you construct a model, analysing that model helps you gain insight into it. Tasks, such as sensitivity analysis, can be added to a model to isolate those components most likely to have an impact on the desired output. Or you can perform parameter estimation to fit values of missing parameters to experimental data. You perform analysis by selecting the relevant task, which is then added to the model browser and is executed by clicking the Run button. You can also create user-defined tasks and include them in models, or share them with peer modellers for re-use.
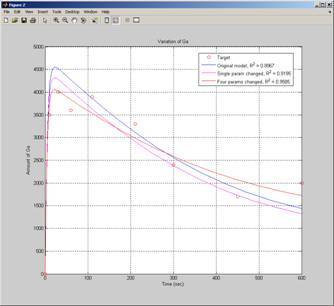
Figure 2: Parameter estimation allows unknown parameters in the model to be fit to external data.
Communicating your results
Modelling can only be effective if the results can be communicated easily and clearly to the relevant audiences. Converting scripts or results into intuitive graphs or models is time-consuming and sometimes ineffective. Results must be in a format and language the intended recipients can decipher.
In SimBiology, you can automatically generate a customisable HTML report by clicking on model component types, such as the graphical layout of the model, graphical output or tables in the SimBiology report generator. You can export graphical images of the model to leverage the visual nature of the model. Images from the model can also be exported as SBML files.
Summary
Modellers at pharmaceutical and biotech companies are under a great a deal of pressure to help alleviate some of the challenges that pharmaceutical researchers and developers are experiencing. The sheer nature of biological research is complex, and trying to create a model to represent biological systems is even more so. Strong modelling capabilities can bring significant value and competitive advantage to a pharmaceutical company. With the right software platform, companies will start to see the increase in efficiency that they are currently seeking.
Interoperability is critical: preparing for the next pandemic now
COVID-19 highlighted the lack of interoperability in our health systems. Here's how ETL...
Transforming health care through digitisation
Australian health regulations require medical records to be retained for decades, with physical...
AI hallucinations are eroding trust in lab tools — but there are solutions
Several key factors will shape whether AI becomes a truly reliable scientific partner or remains...




