RIKEN releases free bioinformatics tool
The RIKEN Center for Life Science Technology has released ZENBU, a freely available bioinformatics tool. The system enables researchers to integrate, visualise and compare large amounts of genomic information resulting from large-scale, next-generation sequencing (NGS) experiments.
NGS protocols such as RNA-seq, ChIP-seq and CAGE have enabled the genome-wide discovery of transcripts and transcription factor binding sites, which is key to understanding the molecular mechanisms underlying cell function in healthy and diseased individuals and the development of diseases like cancer. The integration of data from multiple experiments is an important aspect of the interpretation of results; however, the growing number of datasets makes thorough analysis a cumbersome task.
Writing in the journal Nature Biotechnology, Jessica Severin and colleagues describe the development of ZENBU, a tool that combines a genome browser with data analysis and a linked expression view, to facilitate the interactive visualisation and comparison of results from large numbers of NGS datasets. ZENBU is freely available for use on the web and for installation in individual laboratories, and all ZENBU sites are connected and continuously share data.
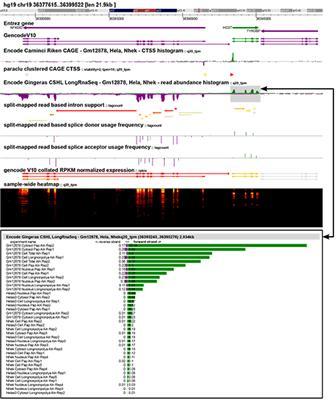
The key difference between ZENBU and previous tools is the ability to dynamically combine thousands of experimental datasets in an interactive visualisation environment through linked genome location and expression signal views. This allows scientists to compare their own experiments against the over 6000 ENCODE and FANTOM consortium datasets currently loaded into the system.
ZENBU allows for novel data exploration through ‘on-demand’ data processing and interactive linked-visualisations and is able to make many views from the same primary sequence alignment data, which users can upload from BAM, BED, GFF and tab-text files. It is designed to integrate millions of experiments/datasets of any kind - RNA-seq, ChIP-seq, CAGE and more.
“By distributing the data and servers, we encourage scientists to load and share their published data to help build a comprehensive resource to further advance research efforts and collaborations around the world,” said the researchers.
The tool can be accessed or downloaded from http://fantom.gsc.riken.jp/zenbu/.
Beckman Coulter Life Sciences announces Automata partnership
Beckman Coulter Life Sciences has announced a strategic partnership with Automata — to...
AI spots hidden objects in chest scans
A new artificial intelligence (AI) tool can spot hard-to-see objects lodged in patients'...
Drying biofluid droplets used to diagnose disease
A newly developed workflow relies on the drying process of biofluid droplets to distinguish...



